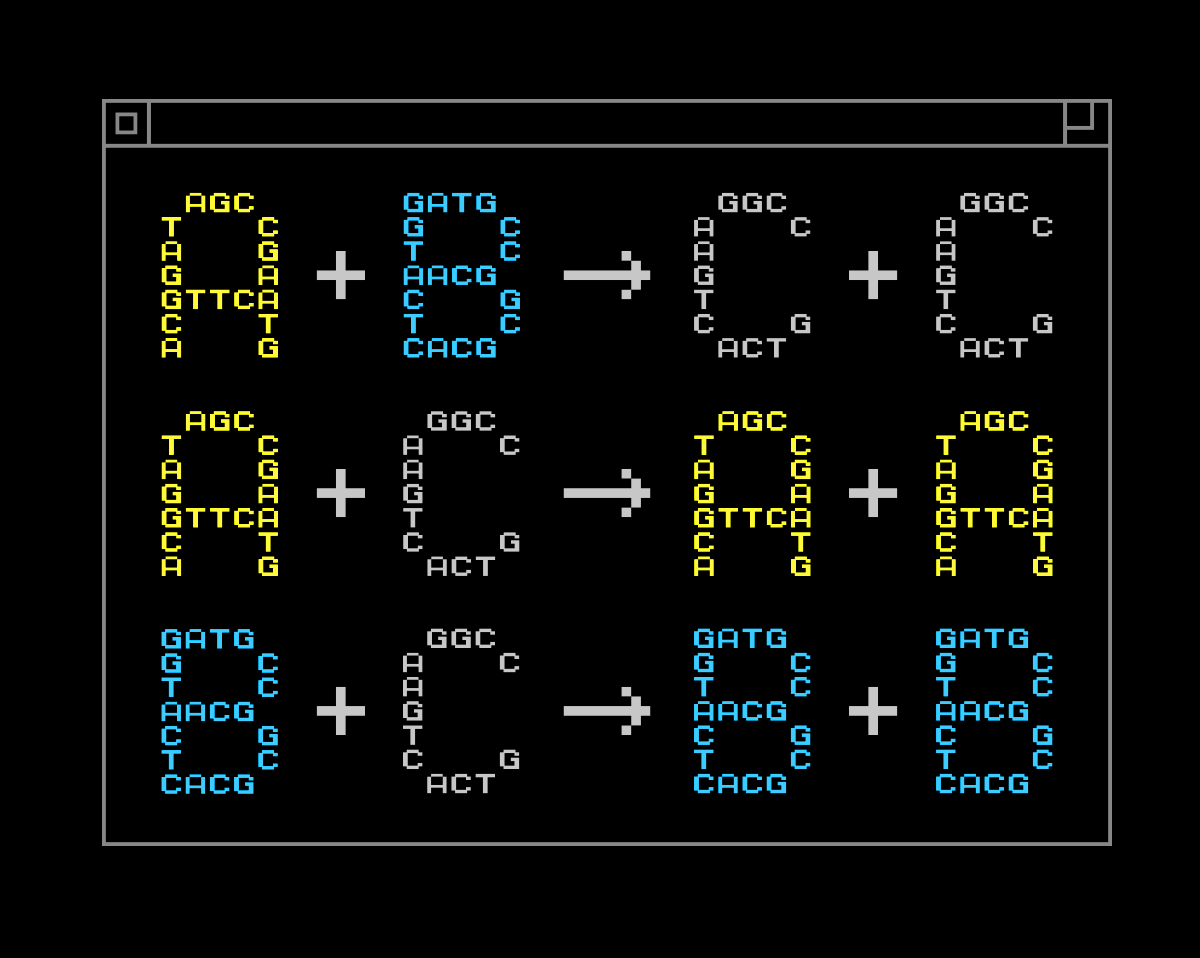Via University of Washington
-----
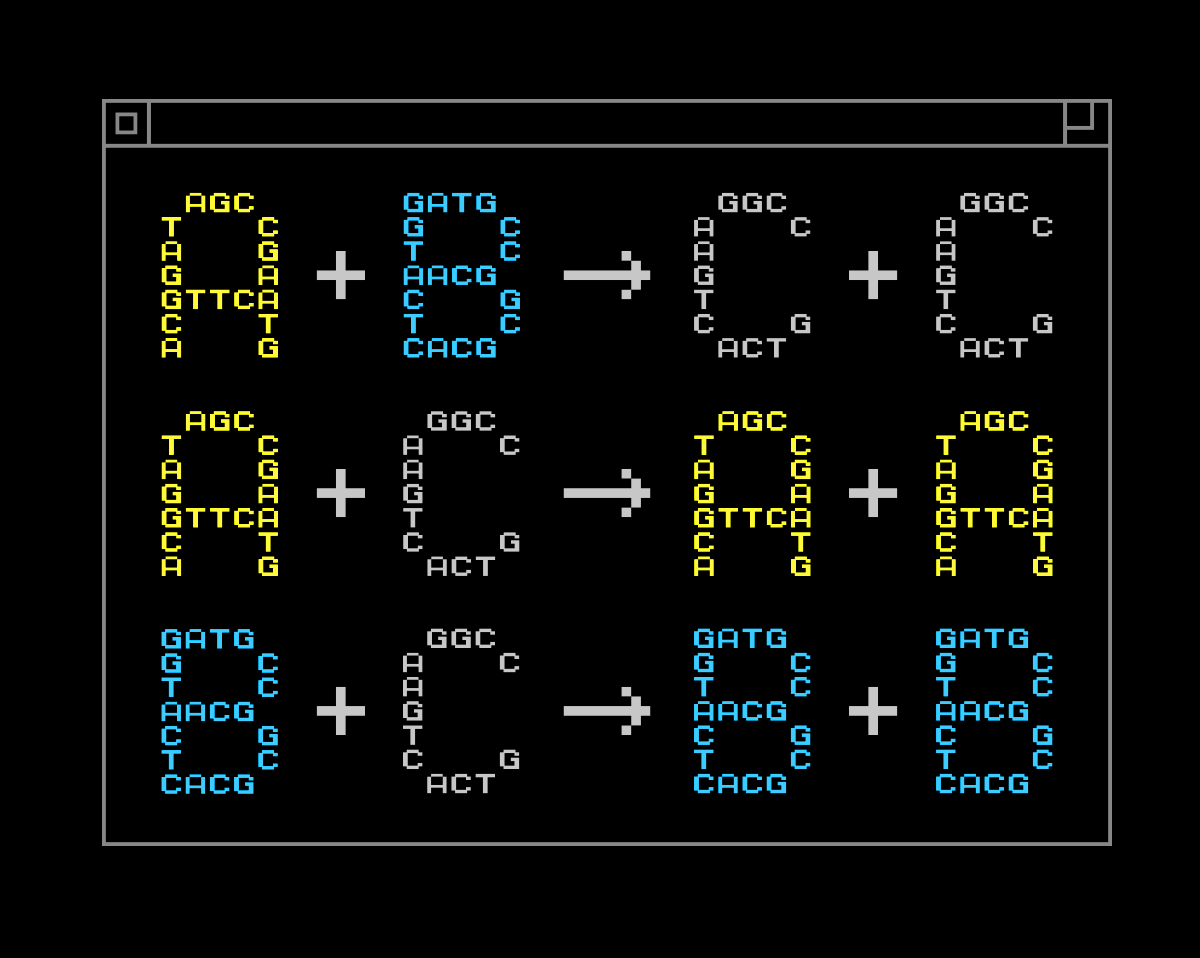
Yan Liang, L2XY2.com
An example of a chemical program. Here, A, B and C are different chemical species.
Similar to using Python or Java to write code for a computer,
chemists soon could be able to use a structured set of instructions to
“program” how DNA molecules interact in a test tube or cell.
A team led by the University of Washington has developed a
programming language for chemistry that it hopes will streamline efforts
to design a network that can guide the behavior of chemical-reaction
mixtures in the same way that embedded electronic controllers guide
cars, robots and other devices. In medicine, such networks could serve
as “smart” drug deliverers or disease detectors at the cellular level.
The findings were published online this week (Sept. 29) in Nature Nanotechnology.
Chemists and educators teach and use chemical reaction networks, a
century-old language of equations that describes how mixtures of
chemicals behave. The UW engineers take this language a step further and
use it to write programs that direct the movement of tailor-made
molecules.
“We start from an abstract, mathematical description of a chemical
system, and then use DNA to build the molecules that realize the desired
dynamics,” said corresponding author Georg Seelig,
a UW assistant professor of electrical engineering and of computer
science and engineering. “The vision is that eventually, you can use
this technology to build general-purpose tools.”
Currently, when a biologist or chemist makes a certain type of
molecular network, the engineering process is complex, cumbersome and
hard to repurpose for building other systems. The UW engineers wanted to
create a framework that gives scientists more flexibility. Seelig
likens this new approach to programming languages that tell a computer
what to do.
“I think this is appealing because it allows you to solve more than
one problem,” Seelig said. “If you want a computer to do something else,
you just reprogram it. This project is very similar in that we can tell
chemistry what to do.”
Humans and other organisms already have complex networks of
nano-sized molecules that help to regulate cells and keep the body in
check. Scientists now are finding ways to design synthetic systems that
behave like biological ones with the hope that synthetic molecules could
support the body’s natural functions. To that end, a system is needed
to create synthetic DNA molecules that vary according to their specific
functions.
The new approach isn’t ready to be applied in the medical field, but
future uses could include using this framework to make molecules that
self-assemble within cells and serve as “smart” sensors. These could be
embedded in a cell, then programmed to detect abnormalities and respond
as needed, perhaps by delivering drugs directly to those cells.
Seelig and colleague Eric Klavins, a UW associate professor of electrical engineering, recently received $2 million
from the National Science Foundation as part of a national initiative
to boost research in molecular programming. The new language will be
used to support that larger initiative, Seelig said.
Co-authors of the paper are Yuan-Jyue Chen, a UW doctoral student in
electrical engineering; David Soloveichik of the University of
California, San Francisco; Niranjan Srinivas at the California Institute
of Technology; and Neil Dalchau, Andrew Phillips and Luca Cardelli of
Microsoft Research.
The research was funded by the National Science Foundation, the
Burroughs Wellcome Fund and the National Centers for Systems Biology.